Description
[[Description::![]() OXPHOS capacity (P) is the respiratory capacity of mitochondria in the ADP-activated state of oxidative phosphorylation, at saturating concentrations of ADP (possibly in contrast to State 3), inorganic phosphate, oxygen, and defined reduced substrates.
» MiPNet article]]
OXPHOS capacity (P) is the respiratory capacity of mitochondria in the ADP-activated state of oxidative phosphorylation, at saturating concentrations of ADP (possibly in contrast to State 3), inorganic phosphate, oxygen, and defined reduced substrates.
» MiPNet article]]
Abbreviation: Has abbr::''P''
Reference: [[Info::Gnaiger 2014 MitoPathways, Gnaiger 2009 Int J Biochem Cell Biol]]
MitoPedia concepts:
Respiratory state,
SUIT concept,
Recommended
MitoPedia methods:
Respirometry
MitoPedia topics:
MitoPedia topic::EAGLE
- » Keywords
- » ADP
- » CS
- » Dyscoupling
- » Flow
- » Flux control ratio
- » Free OXPHOS capacity
- » Gnaiger 2014 MitoPathways
- » Oxidative phosphorylation
- » Oxygen flow
- » Oxygen flux
- » P/E
- » State 3
- » Uncoupling
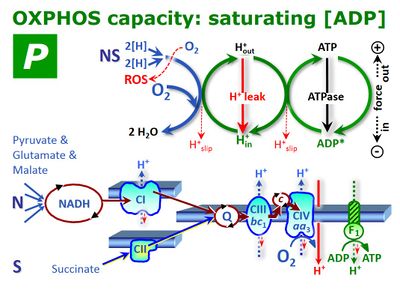
OXPHOS (P) and State 3
| Has title::Gnaiger E (2014) OXPHOS (''P'') and State 3. Mitochondr Physiol Network 2016-07-15. |
Was written by::OROBOROS (Was published in year::2016) Was published in journal::MiPNet
Abstract: [[has abstract::![]() OXPHOS capacity (P) is the respiratory capacity of mitochondria in the ADP-activated state of oxidative phosphorylation, at saturating concentrations of ADP (possibly in contrast to State 3), inorganic phosphate, oxygen, and defined reduced substrates. Since OXPHOS is partially coupled, intrinsic uncoupling and dyscoupling contribute to the control of flux in the OXPHOS state (State P). Oxygen consumption in the OXPHOS state, P, therefore, is partitioned into the free OXPHOS capacity, ≈P, strictly coupled to phosphorylation, ~P, and nonphosphorylating LEAK respiration, LP, compensating for proton leaks, slip and cation cycling: P = ≈P+LP. It is frequently assumed that LEAK respiration, L, as measured in the LEAK state, overestimates the LEAK component of respiration, LP, as measured in the OXPHOS state, particularly if the protonmotive force is not adjusted to equivalent levels in L and LP. However, if the LEAK component increases with enzyme turnover during P, the low enzyme turnover during L may counteract the effect of the higher Δpmt. OXPHOS capacity is expressed (i) per mt-marker (O2 flux per mt-protein, CS, etc); if ETS capacity, E, is used as a functional mitochondrial marker, then OXPHOS capacity is expressed as the P/E ratio (flux control ratio). (ii) OXPHOS capacity is expressed per tissue or cell mass, integrating mt-quantity (density) and mt-quality (O2 flux). (iii) OXPHOS capacity is expressed per cell (O2 flow), which then is a function of mt-density, mt-quality, and cell size. If conditions for measurement and expression of respiration vary, explicit symbols are used, expressing OXPHOS capacity as oxygen flux in state P, JO2P or as oxygen flow in state P, IO2P. If these conditions are defined and remain consistent within a given context, then the simple symbol P for respiratory state can be used to substitute the more explicit expression for respiratory activity.]]
OXPHOS capacity (P) is the respiratory capacity of mitochondria in the ADP-activated state of oxidative phosphorylation, at saturating concentrations of ADP (possibly in contrast to State 3), inorganic phosphate, oxygen, and defined reduced substrates. Since OXPHOS is partially coupled, intrinsic uncoupling and dyscoupling contribute to the control of flux in the OXPHOS state (State P). Oxygen consumption in the OXPHOS state, P, therefore, is partitioned into the free OXPHOS capacity, ≈P, strictly coupled to phosphorylation, ~P, and nonphosphorylating LEAK respiration, LP, compensating for proton leaks, slip and cation cycling: P = ≈P+LP. It is frequently assumed that LEAK respiration, L, as measured in the LEAK state, overestimates the LEAK component of respiration, LP, as measured in the OXPHOS state, particularly if the protonmotive force is not adjusted to equivalent levels in L and LP. However, if the LEAK component increases with enzyme turnover during P, the low enzyme turnover during L may counteract the effect of the higher Δpmt. OXPHOS capacity is expressed (i) per mt-marker (O2 flux per mt-protein, CS, etc); if ETS capacity, E, is used as a functional mitochondrial marker, then OXPHOS capacity is expressed as the P/E ratio (flux control ratio). (ii) OXPHOS capacity is expressed per tissue or cell mass, integrating mt-quantity (density) and mt-quality (O2 flux). (iii) OXPHOS capacity is expressed per cell (O2 flow), which then is a function of mt-density, mt-quality, and cell size. If conditions for measurement and expression of respiration vary, explicit symbols are used, expressing OXPHOS capacity as oxygen flux in state P, JO2P or as oxygen flow in state P, IO2P. If these conditions are defined and remain consistent within a given context, then the simple symbol P for respiratory state can be used to substitute the more explicit expression for respiratory activity.]]
• O2k-Network Lab: Was published by MiPNetLab::AT Innsbruck Gnaiger E
Labels:
Coupling state: Coupling states::OXPHOS
HRR: Instrument and method::Theory
Related terms in Bioblast
 OXPHOS, P
OXPHOS, P
 ROUTINE, R
ROUTINE, R
 ETS, E
ETS, E
 LEAK, L
LEAK, L
 ROX, R
ROX, R
- Free OXPHOS capacity, ≈P = P-L
- Free ROUTINE activity, ≈R = R-L
- Free ETS capacity, ≈E = E-L
- Excess E-P capacity, ExP = E-P
- Excess E-R capacity, ExR = E-R
- » List of publications: OXPHOS
Sort in ascending/descending order by a click on one of the small symbols in squares below. Default sorting: chronological. Empty fields appear first in ascending order.
{{#ask: Coupling states::OXPHOS |?Was published in year=Year |?Has title=Reference |?Coupling states=Coupling |?Pathways=Pathway |?Preparation=Preparation |?Mammal and model=Organism |?Tissue and cell=Tissue;cell |format=broadtable |limit=5000 |offset=0 |sort=Was published in year |order=descending }}
Abstracts: OXPHOS
Sort in ascending/descending order by a click on one of the small symbols in squares below.
{{#ask: Coupling states::OXPHOS |?Was submitted in year=Year |?Has title=Reference |?Coupling states=Coupling |?Pathways=Pathway |?Preparation=Preparation |?Mammal and model=Organism |?Tissue and cell=Tissue;cell |format=broadtable |limit=5000 |offset=0 |sort=Was submitted in year |order=descending }}

