Semantic search
| Term | Abbreviation | Description |
|---|---|---|
| Proton leak | Flux of protons driven by the protonmotive force across the inner mt-membrane, bypassing the ATP synthase and thus contributing to LEAK respiration. Proton leak-flux depends non-linearly (non-ohmic) on the protonmotive force. Compare: Proton slip. | |
| Proton pump | Mitochondrial proton pumps are large enzyme complexes (CI, CIII, CIV, CV) spanning the inner mt-membrane, partially encoded by mtDNA. CI, CIII and CIV are proton pumps that drive protons against the electrochemical protonmotive force, driven by electron transfer from reduced substrates to oxygen. In contrast, ATP synthase (also known as CIV) is a proton pump that utilizes the energy of proton flow along the protonmotive force to drive phosphorylation of ADP to ATP. | |
| Proton slip | Proton slip is a property of the proton pumps (Complexes CI, CIII, and CIV) when the proton slips back to the matrix side within the proton pumping process. Slip is different from the proton leak, which depends on Δp and is a property of the inner mt-membrane (including the boundaries between membrane-spanning proteins and the lipid phase). Slip is an uncoupling process that depends mainly on flux and contributes to a reduction in the biochemical coupling efficiency of ATP production and oxygen consumption. Together with proton leak and cation cycling, proton slip is compensated for by LEAK respiration or LEAK oxygen flux, L. Compare: Proton leak. | |
| Q calibration - DatLab | Q calibration | |
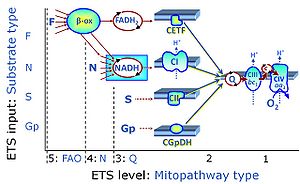
| Q-junction |
The Q-junction is a junction for convergent electron flow in the Electron transfer pathway (ET-pathway) from type N substrates and mt-matrix dehydrogenases through Complex I (CI), from type F substrates and FA oxidation through electron-transferring flavoprotein Complex (CETF), from succinate (S) through Complex II (CII), from glycerophosphate (Gp) through glycerophosphate dehydrogenase Complex (CGpDH), from choline through choline dehydrogenase, from dihydro-orotate through dihydro-orotate dehydrogenase, and other enzyme Complexes into the Q-cycle (ubiquinol/ubiquinone), and further downstream to Complex III (CIII) and Complex IV (CIV). The concept of the Q-junction, with the N-junction and F-junction upstream, provides the rationale for defining Electron-transfer-pathway states and categories of SUIT protocols. | |
| ROUTINE respiration | R | |
| Respiratory acceptor control ratio | RCR | The respiratory acceptor control ratio (RCR) is defined as State 3/State 4 [1]. If State 3 is measured at saturating [ADP], RCR is the inverse of the OXPHOS control ratio, L/P (when State 3 is equivalent to the OXPHOS state, P). RCR is directly but non-linearly related to the P-L control efficiency, jP-L = 1-L/P, with boundaries from 0.0 to 1.0. In contrast, RCR ranges from 1.0 to infinity, which needs to be considered when performing statistical analyses. In living cells, the term RCR has been used for the ratio State 3u/State 4o, i.e. for the inverse L/E ratio [2,3]. Then for conceptual and statistical reasons, RCR should be replaced by the E-L coupling efficiency, 1-L/E [4]. |
| Respiratory chain | RC | The mitochondrial respiratory chain (RC) consists of enzyme complexes arranged to form a metabolic system of convergent pathways for oxidative phosphorylation. In a general sense, the RC includes (1) the electron transfer pathway (ET-pathway), with transporters for the exchange of reduced substrates across the inner mitochondrial membrane, enzymes in the matrix space (particularly dehydrogenases of the tricarboxylic acid cycle), inner membrane-bound electron transfer complexes, and (2) the inner membrane-bound enzymes of the phosphorylation system. |
| Respiratory complexes | CI, CII, CIII, CIV, CETF, CGpDH, .. | Respiratory Complexes are membrane-bound enzymes consisting of several subunits which are involved in energy transduction of the respiratory system. » MiPNet article |
| Respiratory state | Respiratory states of mitochondrial preparations and living cells are defined in the current literature in many ways and with a diversity of terms. Mitochondrial respiratory states must be defined in terms of both, the coupling-control state and the electron-transfer-pathway state. | |
| Respirometry | Respirometry is the quantitative measurement of respiration. Respiration is therefore a combustion, a very slow one to be precise (Lavoisier and Laplace 1783). Thus the basic idea of using calorimetry to explore the sources and dynamics of heat changes were present in the origins of bioenergetics (Gnaiger 1983). Respirometry provides an indirect calorimetric approach to the measurement of metabolic heat changes, by measuring oxygen uptake (and carbon dioxide production and nitrogen excretion in the form of ammonia, urea, or uric acid) and converting the oxygen consumed into an enthalpy change, using the oxycaloric equivalent. Liebig (1842) showed that the substrate of oxidative respiration was protein, carbohydrates, and fat. The sum of these chemical changes of materials under the influence of living cells is known as metabolism (Lusk 1928). The amount (volume STP) of carbon dioxide expired to the amount (volume STP) of oxygen inspired simultaneously is the respiratory quotient, which is 1.0 for the combustion of carbohydrate, but less for lipid and protein. Voit (1901) summarized early respirometric studies carried out by the Munich school on patients and healthy controls, concluding that the metabolism in the body was not proportional to the combustibility of the substances outside the body, but that protein, which burns with difficulty outside, metabolizes with the greatest ease, then carbohydrates, while fats, which readily burns outside, is the most difficultly combustible in the organism. Extending these conclusions on the sources of metabolic heat changes, the corresponding dynamics or respiratory control was summarized (Lusk 1928): The absorption of oxygen does not cause metabolism, but rather the amount of the metabolism determines the amount of oxygen to be absorbed. .. metabolism regulates the respiration. | |
| STPD | STPD | At standard temperature and pressure dry (STPD: 0 °C = 273.15 K and 1 atm = 101.325 kPa = 760 mmHg), the molar volume of an ideal gas, Vm, and Vm,O2 is 22.414 and 22.392 L∙mol-1, respectively. Rounded to three decimal places, both values yield the conversion factor of 0.744 from units used in spiroergometry (VO2max [mL O2·min-1]) to SI units [µmol O2·s-1]. For comparison at normal temperature and pressure dry (NTPD: 20 °C), Vm,O2 is 24.038 L∙mol-1. Note that the SI standard pressure is 100 kPa, which corresponds to the standard molar volume of an ideal gas of 22.711 L∙mol-1 and 22.689 L∙mol-1 for O2. |
| SUIT | SUIT | SUIT is the abbreviation for Substrate-Uncoupler-Inhibitor Titration. SUIT protocols are used with mt-preparations to study respiratory control in a sequence of coupling and pathway control states induced by multiple titrations within a single experimental assay. These studies use biological samples economically to gain maximum information with a minimum amount of cells or tissue. |
| SUIT-001 | RP1 | 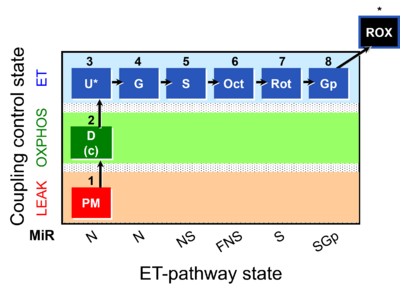 |
| SUIT-001 O2 ce-pce D003 | RP1 ce-pce | 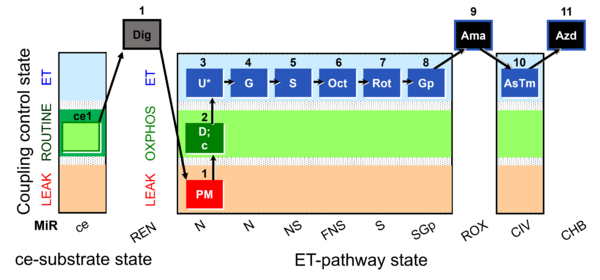 |
| SUIT-001 O2 ce-pce D004 | RP1 ce-pce blood | 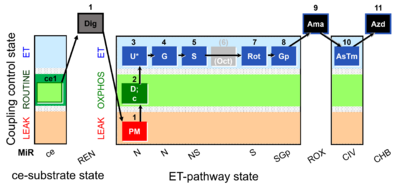 |
| SUIT-001 O2 mt D001 | RP1 mt | 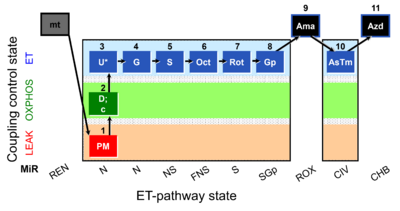 |
| SUIT-001 O2 pfi D002 | RP1 pfi | 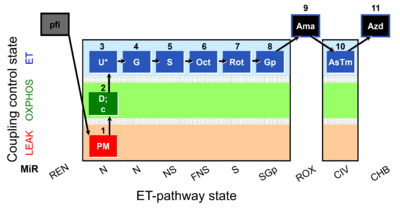 |
| SUIT-002 | RP2 | 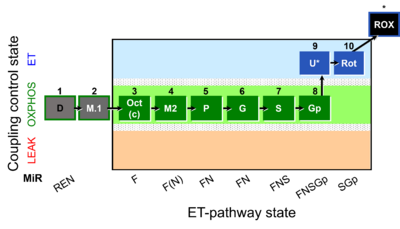 |
| SUIT-002 O2 ce-pce D007 | RP2 ce-pce | 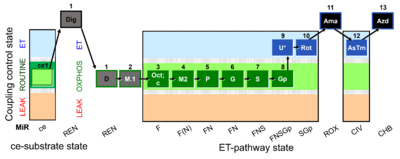 |
| SUIT-002 O2 ce-pce D007a | RP2 ce-pce blood |  |
| SUIT-002 O2 mt D005 | RP2 mt | 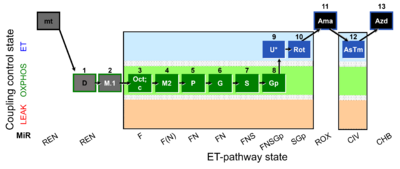 |
| SUIT-002 O2 pfi D006 | RP2 pfi | 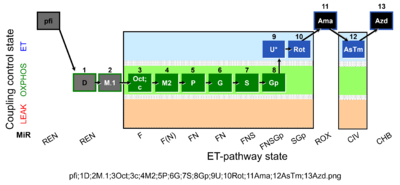 |
| SUIT-003 | CCP-ce | 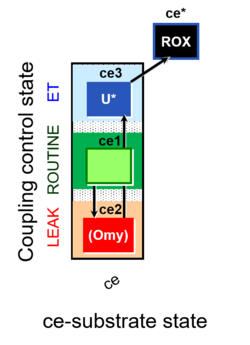 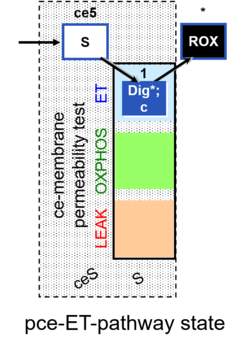 |
| SUIT-003 AmR ce D058 | AmR effect on ce | 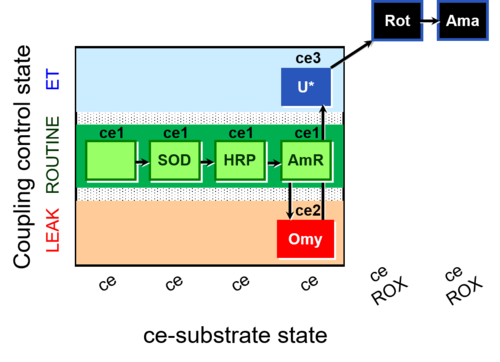 |
| SUIT-003 AmR ce D059 | AmR effect on ce - control | 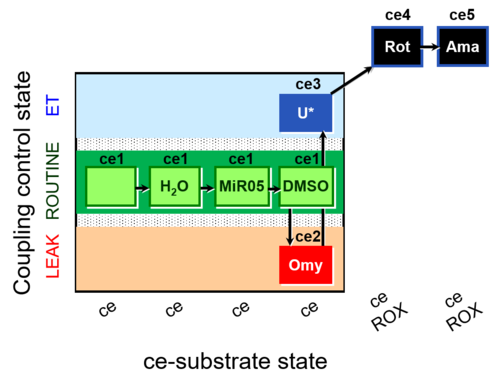 |
| SUIT-003 Ce1;ce1P;ce3U;ce4Glc;ce5M;ce6Rot;ce7S;1Dig;1c;2Ama;3AsTm;4Azd | cePMGlc,S |  |
| SUIT-003 Ce1;ce2U- | ce | 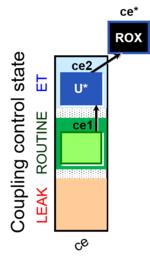 |
| SUIT-003 Ce1;ce3U- | ce | 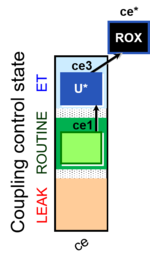 |
| SUIT-003 O2 ce D009 | CCP-ce short | 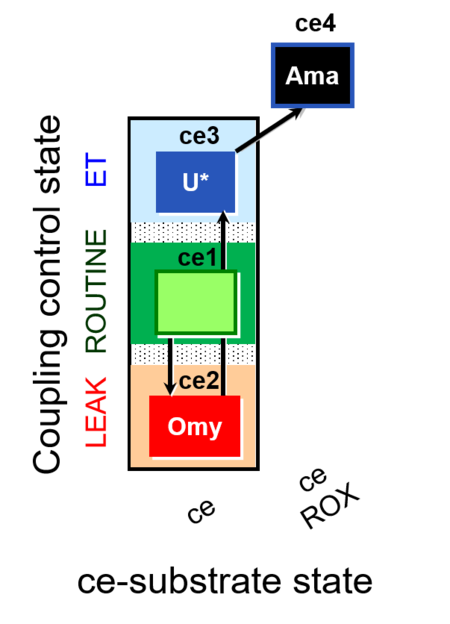 |
| SUIT-003 O2 ce D012 | CCP-ce(P) | 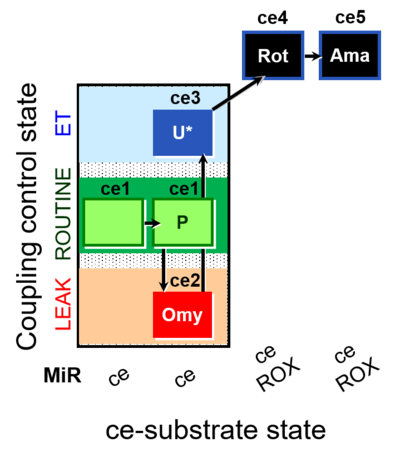 |
| SUIT-003 O2 ce D028 | CCP-ce S permeability test | 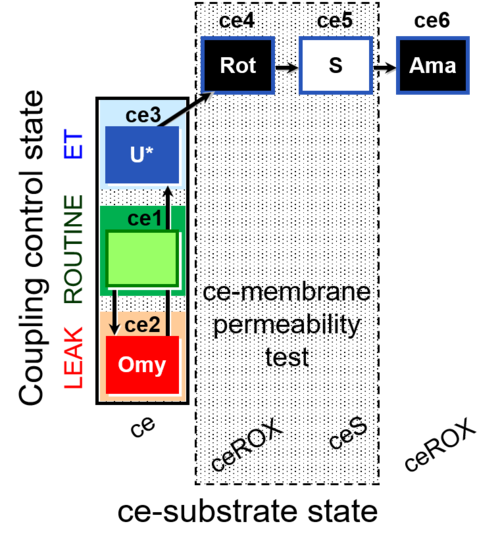 |
| SUIT-003 O2 ce D037 | CCP-ce Crabtree_R | 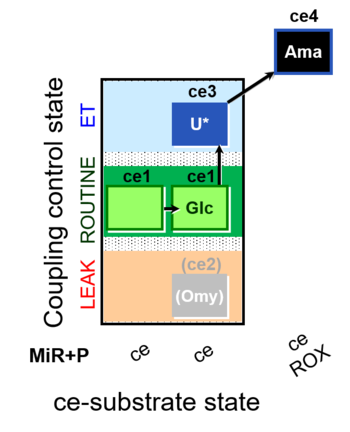 |
| SUIT-003 O2 ce D038 | CCP-ce Crabtree_E | 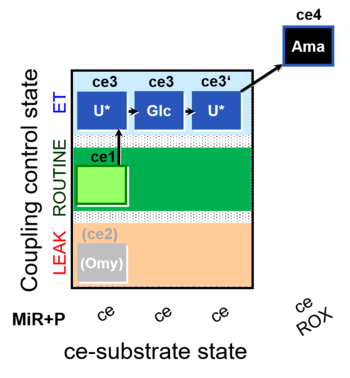 |
| SUIT-003 O2 ce D039 | CCP-ce microalgae | 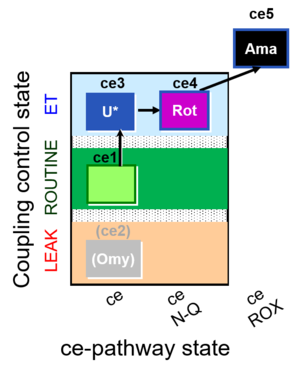 |
| SUIT-003 O2 ce D050 | CCP-ce Snv | 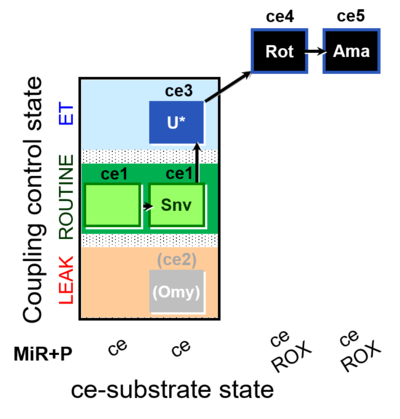 |
| SUIT-003 O2 ce D060 | CCP-ce Snv,Mnanv | 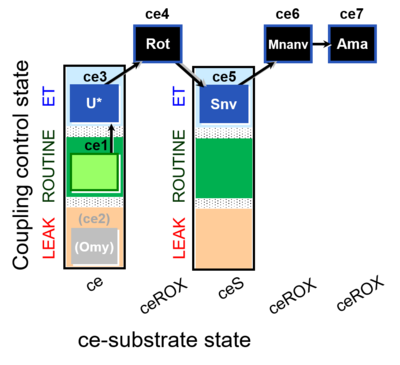 |
| SUIT-003 O2 ce D061 | CCP-ce Snv,Mnanv - control | 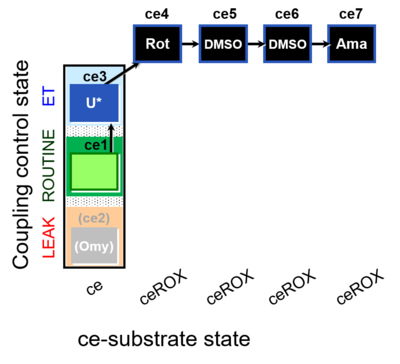 |
| SUIT-003 O2 ce D062 | CCP-ce Snv - control | 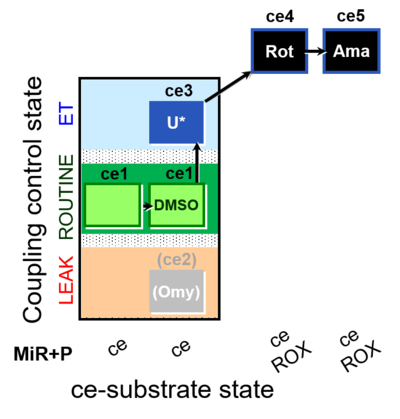 |
| SUIT-003 O2 ce-pce D013 | CCVP-Glc,M | 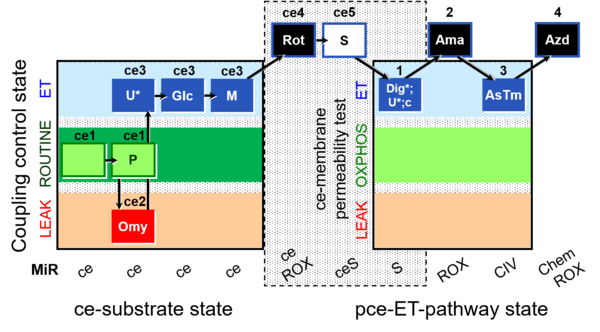 |
| SUIT-003 O2 ce-pce D018 | CCVP-Glc | 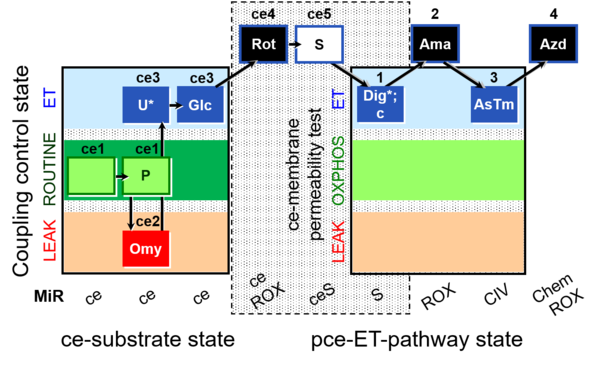 |
| SUIT-003 O2 ce-pce D020 | CCVP | 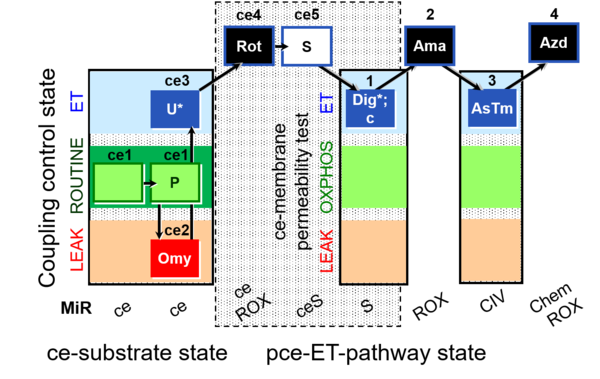 |
| SUIT-003 pH ce D067 | CCP-Crabtree with glycolysis inhibition | 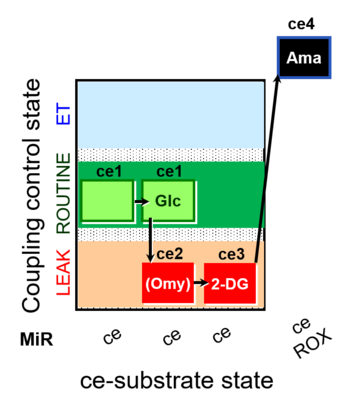 |
| SUIT-004 | RP1-short | 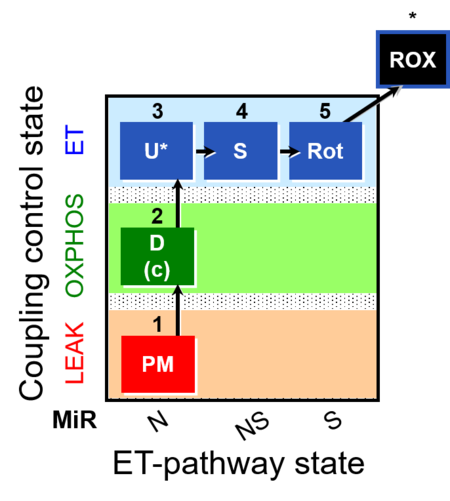 |
| SUIT-004 O2 pfi D010 | RP1-short pfi | 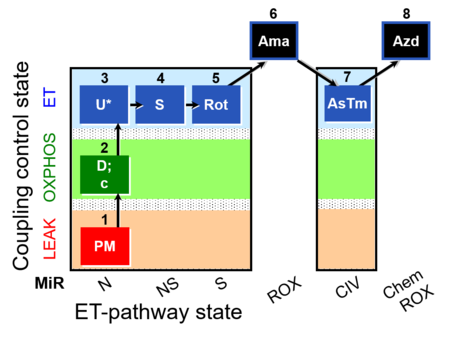 |
| SUIT-005 | RP2-short | 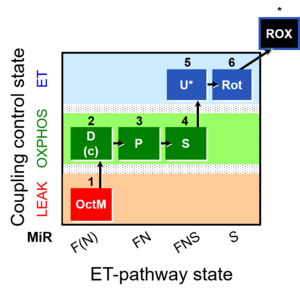 |
| SUIT-005 O2 pfi D011 | RP2-short pfi | 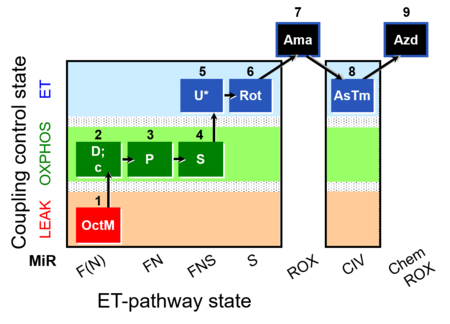 |
| SUIT-006 | CCP-mtprep | 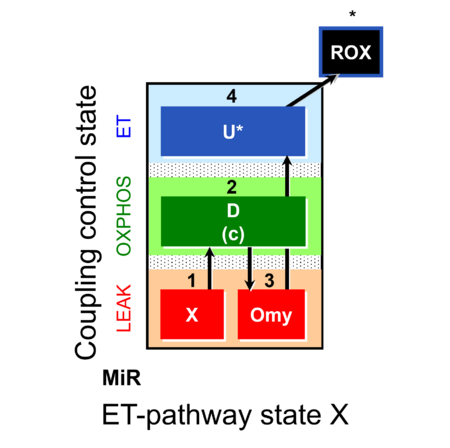 |
| SUIT-006 O2 ce-pce D029 | CCP ce-pce PM | 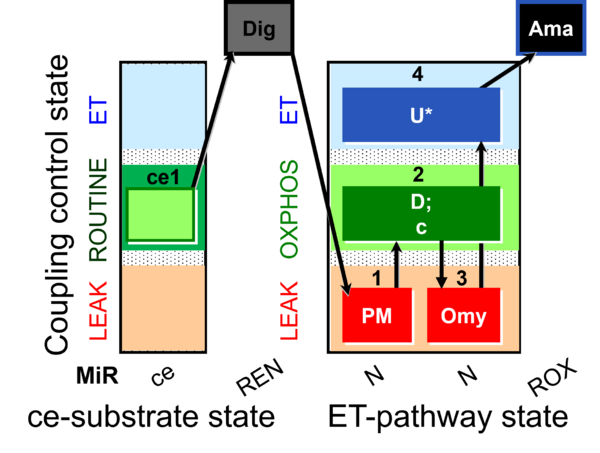 |
| SUIT-006 O2 mt D022 | CCP mt S(Rot) | 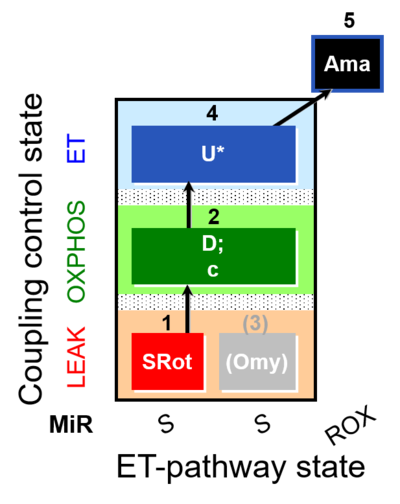 |