Achreja 2022 Nat Metab: Difference between revisions
(Created page with "{{Publication |title=Achreja A, Yu T, Mittal A, Choppara S, Animasahun O, Nenwani M, Wuchu F, Meurs N, Mohan A, Jeon JH, Sarangi I, Jayaraman A, Owen S, Kulkarni R, Cusato M,...") |
No edit summary |
||
| Line 2: | Line 2: | ||
|title=Achreja A, Yu T, Mittal A, Choppara S, Animasahun O, Nenwani M, Wuchu F, Meurs N, Mohan A, Jeon JH, Sarangi I, Jayaraman A, Owen S, Kulkarni R, Cusato M, Weinberg F, Kweon HK, Subramanian C, Wicha MS, Merajver SD, Nagrath S, Cho KR, DiFeo A, Lu X, Nagrath D (2022) Metabolic collateral lethal target identification reveals MTHFD2 paralogue dependency in ovarian cancer. Nat Metab 4:1119-37. https://doi.org/10.1038/s42255-022-00636-3 | |title=Achreja A, Yu T, Mittal A, Choppara S, Animasahun O, Nenwani M, Wuchu F, Meurs N, Mohan A, Jeon JH, Sarangi I, Jayaraman A, Owen S, Kulkarni R, Cusato M, Weinberg F, Kweon HK, Subramanian C, Wicha MS, Merajver SD, Nagrath S, Cho KR, DiFeo A, Lu X, Nagrath D (2022) Metabolic collateral lethal target identification reveals MTHFD2 paralogue dependency in ovarian cancer. Nat Metab 4:1119-37. https://doi.org/10.1038/s42255-022-00636-3 | ||
|info=[https://pubmed.ncbi.nlm.nih.gov/36131208/ PMID: 36131208 Open Access] | |info=[https://pubmed.ncbi.nlm.nih.gov/36131208/ PMID: 36131208 Open Access] | ||
|authors=Achreja A, Yu T, Mittal | |authors=Achreja A, Yu T, Mittal Anjali, Choppara S, Animasahun O, Nenwani M, Wuchu F, Meurs N, Mohan A, Jeon JH, Sarangi I, Jayaraman A, Owen S, Kulkarni R, Cusato M, Weinberg F, Kweon HK, Subramanian C, Wicha MS, Merajver SD, Nagrath S, Cho KR, DiFeo A, Lu X, Nagrath D | ||
|year=2022 | |year=2022 | ||
|journal=Nat Metab | |journal=Nat Metab | ||
| Line 8: | Line 8: | ||
|editor=Gnaiger E | |editor=Gnaiger E | ||
}} | }} | ||
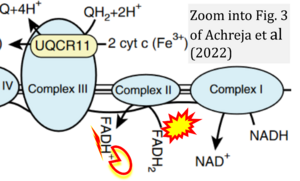
[[File:Achreja 2022 Nat Metab CORRECTION.png|right|300px]] | |||
{{Template:Correction FADH2 and S-pathway}} | |||
{{Labeling | {{Labeling | ||
|diseases=Cancer | |diseases=Cancer | ||
|enzymes=Complex II;succinate dehydrogenase | |enzymes=Complex II;succinate dehydrogenase | ||
}} | }} | ||
Latest revision as of 06:01, 4 September 2023
| Achreja A, Yu T, Mittal A, Choppara S, Animasahun O, Nenwani M, Wuchu F, Meurs N, Mohan A, Jeon JH, Sarangi I, Jayaraman A, Owen S, Kulkarni R, Cusato M, Weinberg F, Kweon HK, Subramanian C, Wicha MS, Merajver SD, Nagrath S, Cho KR, DiFeo A, Lu X, Nagrath D (2022) Metabolic collateral lethal target identification reveals MTHFD2 paralogue dependency in ovarian cancer. Nat Metab 4:1119-37. https://doi.org/10.1038/s42255-022-00636-3 |
Achreja A, Yu T, Mittal Anjali, Choppara S, Animasahun O, Nenwani M, Wuchu F, Meurs N, Mohan A, Jeon JH, Sarangi I, Jayaraman A, Owen S, Kulkarni R, Cusato M, Weinberg F, Kweon HK, Subramanian C, Wicha MS, Merajver SD, Nagrath S, Cho KR, DiFeo A, Lu X, Nagrath D (2022) Nat Metab
Abstract: Recurrent loss-of-function deletions cause frequent inactivation of tumour suppressor genes but often also involve the collateral deletion of essential genes in chromosomal proximity, engendering dependence on paralogues that maintain similar function. Although these paralogues are attractive anticancer targets, no methodology exists to uncover such collateral lethal genes. Here we report a framework for collateral lethal gene identification via metabolic fluxes, CLIM, and use it to reveal MTHFD2 as a collateral lethal gene in UQCR11-deleted ovarian tumours. We show that MTHFD2 has a non-canonical oxidative function to provide mitochondrial NAD+, and demonstrate the regulation of systemic metabolic activity by the paralogue metabolic pathway maintaining metabolic flux compensation. This UQCR11-MTHFD2 collateral lethality is confirmed in vivo, with MTHFD2 inhibition leading to complete remission of UQCR11-deleted ovarian tumours. Using CLIM's machine learning and genome-scale metabolic flux analysis, we elucidate the broad efficacy of targeting MTHFD2 despite distinct cancer genetic profiles co-occurring with UQCR11 deletion and irrespective of stromal compositions of tumours.
• Bioblast editor: Gnaiger E
Correction: FADH2 and Complex II
- FADH2 is shown as the substrate feeding electrons into Complex II (CII). This is wrong and requires correction - for details see Gnaiger (2024).
- Gnaiger E (2024) Complex II ambiguities ― FADH2 in the electron transfer system. J Biol Chem 300:105470. https://doi.org/10.1016/j.jbc.2023.105470 - »Bioblast link«
Labels: Pathology: Cancer
Enzyme: Complex II;succinate dehydrogenase