From Bioblast
Description
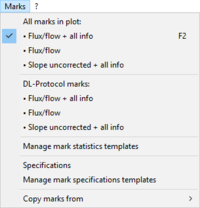
The function Mark specifications is largely replaced by SUIT DL-Protocols and Instrumental DL-Protocols in DatLab 7.4. Mark specifications allow the user to rename Marks in the active plot and save/recall the settings. Rename marks individually by clicking into the horizontal bar, or use corresponding templates for renaming the entire sequence of marks.
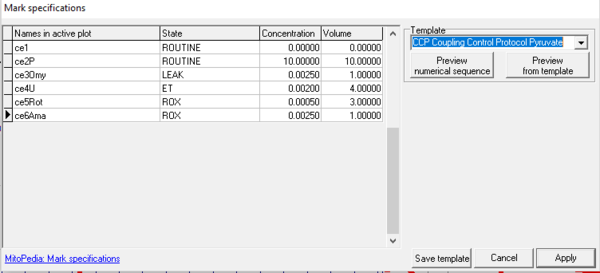
- Mark specifications (name, state, concentration and volume) are shown from the active plot. Define the template name and save template.
- Select a Template, left click on preview from template, and rename all marks of the plot, including the state, concentration and volume.
Reference mark names in DatLab 7
| Reference mark names | Description | SUIT protocol | Reference |
|---|---|---|---|
| MiPNet08.09_IntactCells | Coupling control protocol (ceCCP) in intact cells. | 1ce;2ceOmy;3ceU- | MiPNet08.09 CellRespiration
|
| MiPNet10.04_IntactCells | Coupling control protocol (ceCCP) in intact cells using TIP2k for automatic uncoupler titrations. | 1ce;2ceOmy;3ceU- | MiPNet10.04 CellRespiration
|
| MiPNet21.06 RP1pce | |||
| MiPNet21.06 RP2pce | |||
| MiPNet21.06 RP1mt | |||
| MiPNet21.06 RP2mt |
- » Library of SUIT protocols MitoPedia: SUIT protocols
Keywords
- Bioblast links: Marks in DatLab - >>>>>>> - Click on [Expand] or [Collapse] - >>>>>>>
- Specific
- O2k-Procedures
- MiPNet O2k-Procedures
- » MiPNet26.06 DatLab 7: Guide - Section on setting Marks
- MiPNet O2k-Procedures
- General
MitoPedia O2k and high-resolution respirometry:
DatLab